
n is chosen based on the above two steps. extractSignatures - uses non-negative matrix factorization to decompose the matrix into n signatures.
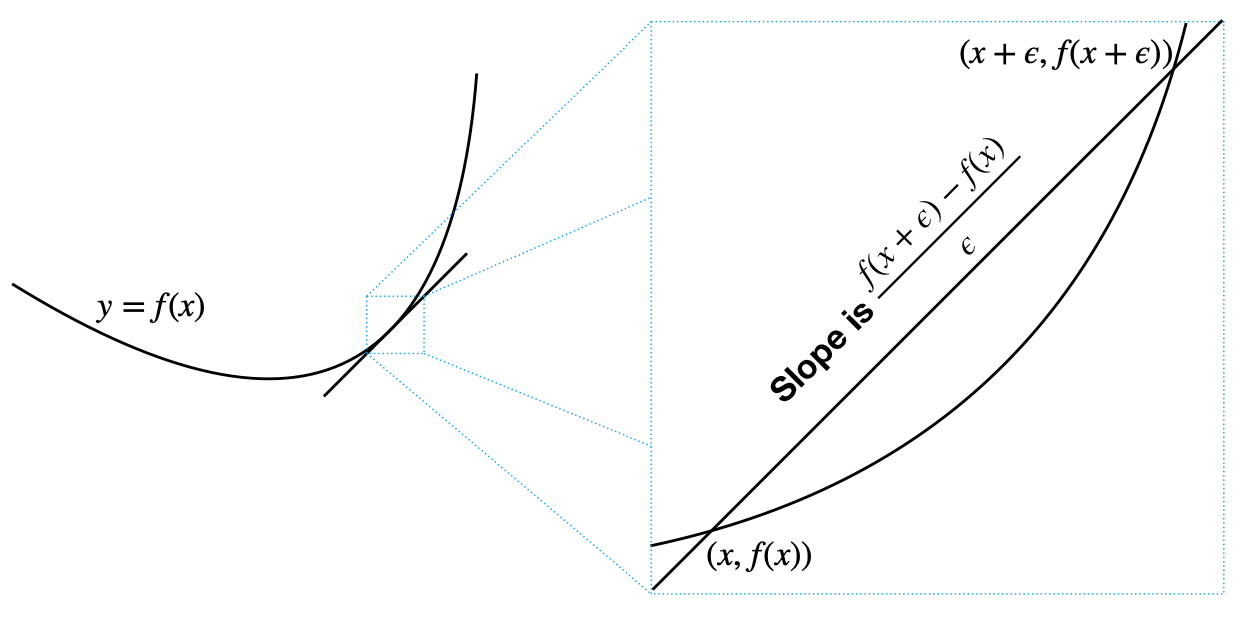
Best possible signature is the value at which Cophenetic correlation drops significantly.


# Hugo_Symbol Primary Relapse pval or ci.up ci.low pt.vs.rt <- mafCompare( m1 = primary.apl, m2 = relapse.apl, m1Name = 'Primary', m2Name = 'Relapse', minMut = 5) print(pt.vs.rt) # $results #Considering only genes which are mutated in at-least in 5 samples in one of the cohort to avoid bias due to genes mutated in single sample. We can plot the results using plotOncodrive. # AlteredSamples clusters muts_in_clusters clusterScores protLen zscore # Missense_Mutation Nonsense_Mutation Splice_Site total MutatedSamples Head(laml.sig) # Hugo_Symbol Frame_Shift_Del Frame_Shift_Ins In_Frame_Del In_Frame_Ins # gene1 gene2 pValue oddsRatio 00 11 01 10 Event somaticInteractions( maf = laml, top = 25, pvalue = c( 0.05, 0.1)) #exclusive/co-occurance event analysis on top 10 mutated genes.


 0 kommentar(er)
0 kommentar(er)
